Ooh, nice one. That’s looking like a winner:
Yeah, I think folks are focusing on phenotype difference instead of genotype differences, which the OP explicitly stated he or she was interested in (the latter, that is).
No, that’s backwards. Corn is an example of gross morphological changes which did not require gross genetic changes.
“corn” doesn’t just mean maize (although it might to the OP).
It’s MrDibble, one word, no “r”.
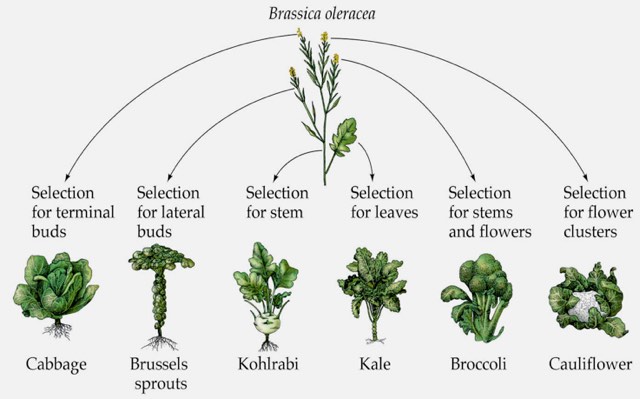
Here is a nice trivia tidbit vaguely related to this discussion and to the subject of substantial phenotype changes among the various Brassica species…
What do you get if you cross cabbage (Brassica oleracea) and rape (Brassica campestris as in rape seed oil)? Seriously… something resembling a turnip.
Plants can make huge morphological changes through adding one or more complete copies of the parental genome. Perhaps a third of all flowering plants exhibit polyploidy of this sort.
Polyploidism is surprisingly easy to achieve. It happens naturally fairly often in plants. It’s really not that big of a deal, achievement-wise.
Also, in many cases, polyploidism itself doesn’t make a big change in the plant. But it is the tool that can be used to make changes. Those extra chromosomes allow a wider range of changes that can be bred for. It’s what you do afterwards that’s the key.
The breeders were trying to produce a version that tasted good enough for humans to eat! Someday they might.
Where have you been? They succeeded years ago.
I beg to differ.
Many “seedless” version of fruits are produced by crossing a diploid with a tetraploid version of the same species - the triploid offspring can undergo normal cell division, but the meiotic cell division needed to produce viable seeds is no longer possible.
It may well be disease resistant wheat , hybridised in the 20th century with very distant relatives in order to transfer in disease resistance from that relative.
Rather than the result occuring in the first or second generation, the results only bear fruit , or seeds in this case, after many generations… See Introgression - Wikipedia
Starlink wheat was GM’ed by gene splicing.
Ploidy is far from the only way to evaluate “gross genetic differences”.
Obviously - I wouldn’t hold up monospecific polyploidy as being particularly indicative. But multispecific polyploidy, I would.
I am told that cherries and strawberries used to be smaller and tastier than they are today, and that a tomato from 70 years ago was leaps and bounds more delicious than the modern one.
Keep in mind, most of those reports are from the elderly whose sense of taste has greatly diminished from their youth and memories of taste are tricky anyway.
What you need is a real comparison between true heirloom fruits and the current ones sold in mass.
For anyone who’s still confused:
These are not steps in a single, complex transformation. Rather they are each an example of what happens if you breed the original (wild mustard) in a specific direction. Here’s a good illustration:
This does not show all the outcomes which are now commercial products either.
If the question were which original plant has resulted in the biggest variety of modern food crops, I’m pretty sure wild mustard would win hands down.
But back to the OP, what about some of the Winter-hardy plants, like hardy kiwifruit or pears? They’ve had to go the farthest in very different traits in order to be useful. I’d think that increases the diversity of change. . .
I have some vague recollection of domesticated brassicas originating from some distinct wild types, but I’m going to demand a cite from myself on that one.
This is an American board.
It may seem like there should be some natural quantitative way to measure degree of genetic difference, but I can’t see any obvious way to deal with either polyploidy or hybridization in such a measure. On the one hand, when you’ve got an octoploid crop, three quarters of its genome wasn’t present in the original population. That looks like a big difference. But on the other hand, if you look at the wild type, its entire genome is contained in the crop version’s genome, and so they look almost identical. And then there’s the process: Polyploidy happens fairly easily among plants, and someone else starting with the same wild plant could double it twice and end up with the same octoploid final result.
Similar questions arise with hybrids. Compare a hybrid to either of its direct ancestors, and you find that half of its genome is different. But you’ll get basically the same result every time you hybridize those two plants-- Is that really such a big change?
On the other hand, you can also human-breed a crop that differs in just one single gene, or even just one single base-pair, from the wild type. But which gene or base pair? If you try again to change just one single gene, you’re likely to get something completely different from the first time, and quite possibly nonviable.
I think the best approach here would need to come from information theory. The more information is required to describe a crop’s genetic differences from the wild type, the more different it should be regarded as.
Tomato* leaves* are indeed, slightly poisonous. So is the green unripe fruit.